Data specifications¶
Here we describe how data has to be
- ... organized: data processing can be done for a batch of images, but for this data has to be organized as specified below.
- ...named: FISH-quant extracts important information from the file-names. This requires a certain naming convention described below.
Demo data¶
Already processed demo data (in 3D) can be downloaded from Dropbox. With these data, you can verify if the different analysis steps are properly executed.
A smaller (2D) data-set is directly available from within the FISH-quant interface, when you press on the button Get zipped (demo) data. You can also download it from Dropbox
Organization¶
We strongly recommend the following data-organization on which this workflow has been tested.
The same data organization is also used in the package to perform cell and nuclear segmentation. This permits to easily combine these workflows.
- Images are store as single-channel multi-z-stack tif files, e.g on tif per position and channel. If your data are not single-channel, see the section on how to split channels with Fiji.
- All raw 3D images are stored in a folder
acquisition - All analysis results are stored in subfolder
analysis, where each analysis step has a separate subfolder. This folder can be automatically created as explained in the section about data import.
The organization of the provided test data is the following
├─ fq-imjoy-demo/
│ ├─ acquisition # Folder with raw images
│ │ ├─ test_pos001_cy3.tif
│ │ ├─ test_pos001_dapi.tif
│ │ ├─ test_pos002_cy3.tif
│ │ ├─ test_pos002_dapi.tif
│ ├─ analysis # Folder with all analysis results
│ │ ├─ segmentation-input # Folder with input for segmentation
│ │ │ ├─ ....
│ │ ├─ segmentation-results # Folder with segmentation results
│ │ │ ├─ ....
│ │ ├─ spot-detection # FQ detection results
│ │ │ ├─ test_pos001_cy3__settings.json # Settings files
│ │ │ ├─ test_pos001_cy3__spots.csv # Spot detection results
Naming convention & regular expression¶
FISH-quant will extract important information about the images, such as the field of view or the imaged channel, directly from the file-names. For this, we use a regular expression. A regular expression is used specifies search patterns in text. In our case, we use such an expression to analyze the image file-name and extract important information about the image, such as what channel it is.
Regular expression look a bit complicated, but they provide a lot of flexibility and allow us to not impose a strict naming scheme, but extract the relevant information with these expressions and thus process images with with different naming conventions.
The filenames MUST contain an identifier for:
- experiment (
file_ident): string specifying an experiment, e.g. what gene was imaged. If multiple channel are imaged, this string has to be identical for all images. - field of view (
fov): what position was acquired. As above, has to be identical for multi-channel images. - channel (
channel): which channel was imaged. - image extension (
img_ext): image extension, e.g.tif. Only images with this extension will be loaded.
These informations are extracted by using regular expression and capture groups. Here, we only explain some of the basic concepts, there are many excellent ressources on the web to learn and test your own regular expression, such as https://regex101.com/.
As an example, we use the default naming scheme from the examples above. Here, the different identifies are separated by _ and ordered as <file_ident>_<fov>_<channel>.<img_ext>:
fov img_ext
| |
test_pos001_cy3.tif
| |
file_ident channel
We can now define a regular expression to analyze such a string, and extract the different subgroups. For this, you essentially write a string with fixed elements, and placeholders (tokens) that match you file-name. In order to analyze this file-name, we only need the following elements
.matches any character (except for line terminators)*matches the previous token between zero and unlimited times.\.matches the character.once._matches the character_once.- (?P
.*) is a named capture group (name fov), which allows to extract the part that matches to this subgroup.
With this, we can write the following regular expression
(?P<file_ident>.*)_(?P<fov>.*)_(?P<channel>.*)\.(?P<img_ext>.*)
This allows to analyze a given filename, e.g. test_pos001_cy3.tif and extract the different subgroups
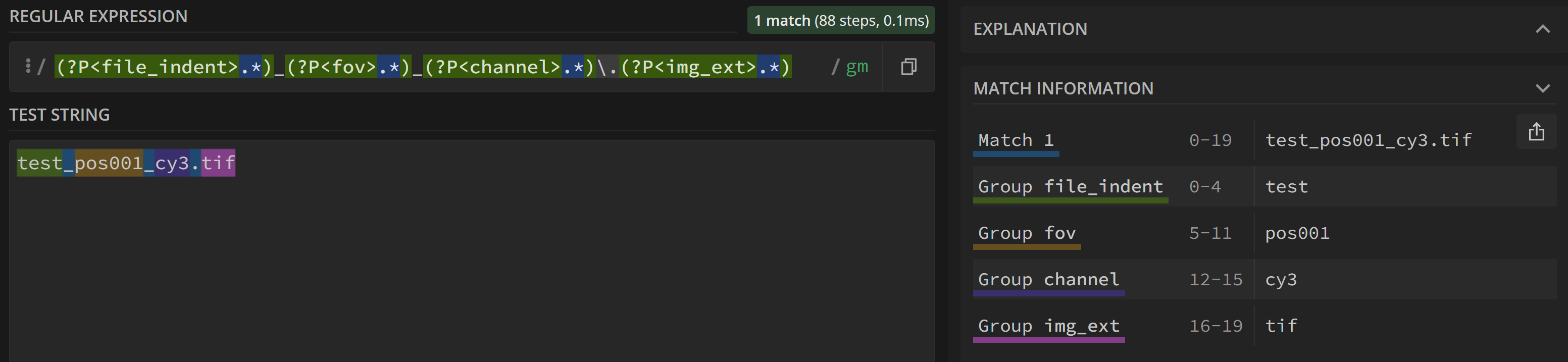
In this example, the fov capture group corresponds to pos001'. If you would like to extract only the number and not
the stringpos, you could explicitly add this string to the regular expression outside of the capture group (_pos(?P
(?P<file_indent>.*)_pos(?P<fov>.*)_(?P<channel>.*)\.(?P<img_ext>.*)
Now, the capture group only contains 001. To get a better feeling for this, you can also play around
on https://regex101.com/).
IMPORTANT: if you have multiple images, e.g. different channels, of the same field of view, and you want to load them at the same time into the interfacem you have to guarantee that the fov and file_ident are identical for these images. If they are not, the images will not be matched, and the images will not show up. Construct your regular expression accordingly!